Mixture model example for three sided heterogeniety
18 May, 2021
Source:vignettes/acblm_example.Rmd
acblm_example.RmdSimulating and estimating the mixture model
require(acblm)
require(knitr)
require(kableExtra)
options(knitr.table.format = "html")
knitr::opts_chunk$set(dev = "png", dev.args = list(type = "cairo-png"))Simulating a data set
In time period t, the distribution which depends on the type αi (worker in paper), class mit (manager in paper) and the class kit (firm class in paper).
Pr[Yit≤y|mit=m,kit=k,αi=α]=Fmkα(y)
set.seed(3236)
# three sided model
model_test = m2.mixt.new(nk=2,nf=2,nb=2)
# model initializer for beta tests
model <- ModelInitializer()
# assign the means of distributions system has simple complementarity
model_test$A1[1,,]=model$A1[1,1:2,]
model_test$A1[2,,]=model$A1[1,3:4,]
# assign the variance of distribution ( small: will help in convergence on desktop)
model_test$S1[] = 0.1
model_test$S2[] = 0.1
# Simple case when distribution at T=2 is same to T=1
model_test$A2 = model_test$A1
model_test$NNm[,,,] = 1000
model_test$pk1[,,]=0.5
model_test$pk0[,,]=0.5
pk1 = rdirichlet(2*2*2*2,rep(1,2))
dim(pk1) = c(2*2, 2*2 , 2)
model_test$pk1 = pk1
pk0 = rdirichlet(2*2,rep(1,2))
dim(pk0) = c(2,2,2)
model_test$pk0 = pk0
# simulate the data
test_data <- Simulate.data.threeSided(model_test)Set the model controls
ctrl <- set.solver.controls(model_test, # Model
n_startValues=1, # number of starting values
stayers_sample=0.1) # if want to subsample the stayers data Step 1 of estimation : Clustering manager and firms
ad_employee_em <- threeSided.Clustering(test_data) # simulated data ## INFO [2021-05-18 13:03:28] here i am
## INFO [2021-05-18 13:03:28] generating measures for firms
## INFO [2021-05-18 13:03:28] processing 6000 unique id's
## INFO [2021-05-18 13:03:28] computing measures...
## INFO [2021-05-18 13:03:29] computing weights...
## INFO [2021-05-18 13:03:29] clustering T=51.002687, Nw=20 , measure=ecdf
## INFO [2021-05-18 13:03:29] running weigthed kmeans step=250 total=1000
## INFO [2021-05-18 13:03:29] nobs=6000 nmeasures=20
## INFO [2021-05-18 13:03:30] running weighted k-means [25%] completed
## INFO [2021-05-18 13:03:30] running weighted k-means [50%] completed <<<<<
## INFO [2021-05-18 13:03:31] running weighted k-means [75%] completed <<<<<
## INFO [2021-05-18 13:03:32] running weighted k-means [100%] completed <<<<<
## INFO [2021-05-18 13:03:32] k=2 WSS=945.565383 nstart=1000 ids=6000
## INFO [2021-05-18 13:03:32] generating measures for managers
## INFO [2021-05-18 13:03:32] processing 6000 unique id's
## INFO [2021-05-18 13:03:32] computing measures...
## INFO [2021-05-18 13:03:32] computing weights...
## INFO [2021-05-18 13:03:33] clustering T=51.008960, Nw=20 , measure=ecdf
## INFO [2021-05-18 13:03:33] running weigthed kmeans step=250 total=1000
## INFO [2021-05-18 13:03:33] nobs=6000 nmeasures=20
## INFO [2021-05-18 13:03:33] running weighted k-means [25%] completed
## INFO [2021-05-18 13:03:34] running weighted k-means [50%] completed <<<<<
## INFO [2021-05-18 13:03:35] running weighted k-means [75%] completed <<<<<
## INFO [2021-05-18 13:03:36] running weighted k-means [100%] completed <<<<<
## INFO [2021-05-18 13:03:36] k=2 WSS=945.607584 nstart=1000 ids=6000Step 2 of estimation : Model parameters estimation (modified EM: see paper)
my_model_test_cluster <- estimation.threeSided.model(model_test, # model
ad_employee_em, # data with step 1 estimation results
ctrl) # control parametrs for solver ## Warning: 'cBind' is deprecated.
## Since R version 3.2.0, base's cbind() should work fine with S4 objects## Warning: 'rBind' is deprecated.
## Since R version 3.2.0, base's rbind() should work fine with S4 objects## INFO [2021-05-18 13:03:41] [ 29][para0][final] lik=-817.8422 dlik=5.8178e-08 liks=-8.1752e+02 likm=0.0000e+00
## INFO [2021-05-18 13:03:41] res para : value at model start
## INFO [2021-05-18 13:03:41] starting repetitions with 1 nodes
## INFO [2021-05-18 13:03:43] [ 37][paraf (1/1)][final] lik=-95979.7050 dlik=8.9607e-08 liks=-9.5978e+04 likm=0.0000e+00
## INFO [2021-05-18 13:03:48] [ 62][para1 (1/1)][final] lik=-138.3642 dlik=9.8202e-08 liks=-1.3805e+02 likm=0.0000e+00
## INFO [2021-05-18 13:03:50] [ 22][move1 (1/1)][final] lik=11630.2041 dlik=7.2779e-08 liks=1.1631e+04 likm=0.0000e+00
## INFO [2021-05-18 13:03:50] done with reptitions 1/1
## INFO [2021-05-18 13:03:50] drawing 0.100000 from the stayers
## INFO [2021-05-18 13:03:50] selecting best model
## INFO [2021-05-18 13:03:50] print pk0,nf*nx,nk
## [,1] [,2]
## [1,] 0.7773345 0.2226655
## [2,] 0.8004161 0.1995839
## [3,] 0.2289786 0.7710214
## [4,] 0.4192576 0.5807424
## , , 1
##
## [,1] [,2] [,3] [,4]
## [1,] 0.1521758 0.3009681 0.1736538 0.3569221
##
## , , 2
##
## [,1] [,2] [,3] [,4]
## [1,] 0.8478242 0.6990319 0.8263462 0.6430779
##
## INFO [2021-05-18 13:03:51] drawing here
## cor_kl cor_km cor_lm cov_kl cov_km cov_lm var_k var_l var_m rsq
## 1 0.183 0.0464 0.0104 0.0574 0.0282 0.0012 0.7599 0.0324 0.1208 0.7002# Estimation results for mean
my_model_test_cluster$model$A1[1,,]## [,1] [,2]
## [1,] 0.1471967 0.02025321
## [2,] 0.4167360 0.05339750
## [3,] 0.7037331 0.06984640
## [4,] 0.9861827 0.10224342Proportion plots
# for manager class 1
threeSided.proportion.plot(my_model_test_cluster, m=1)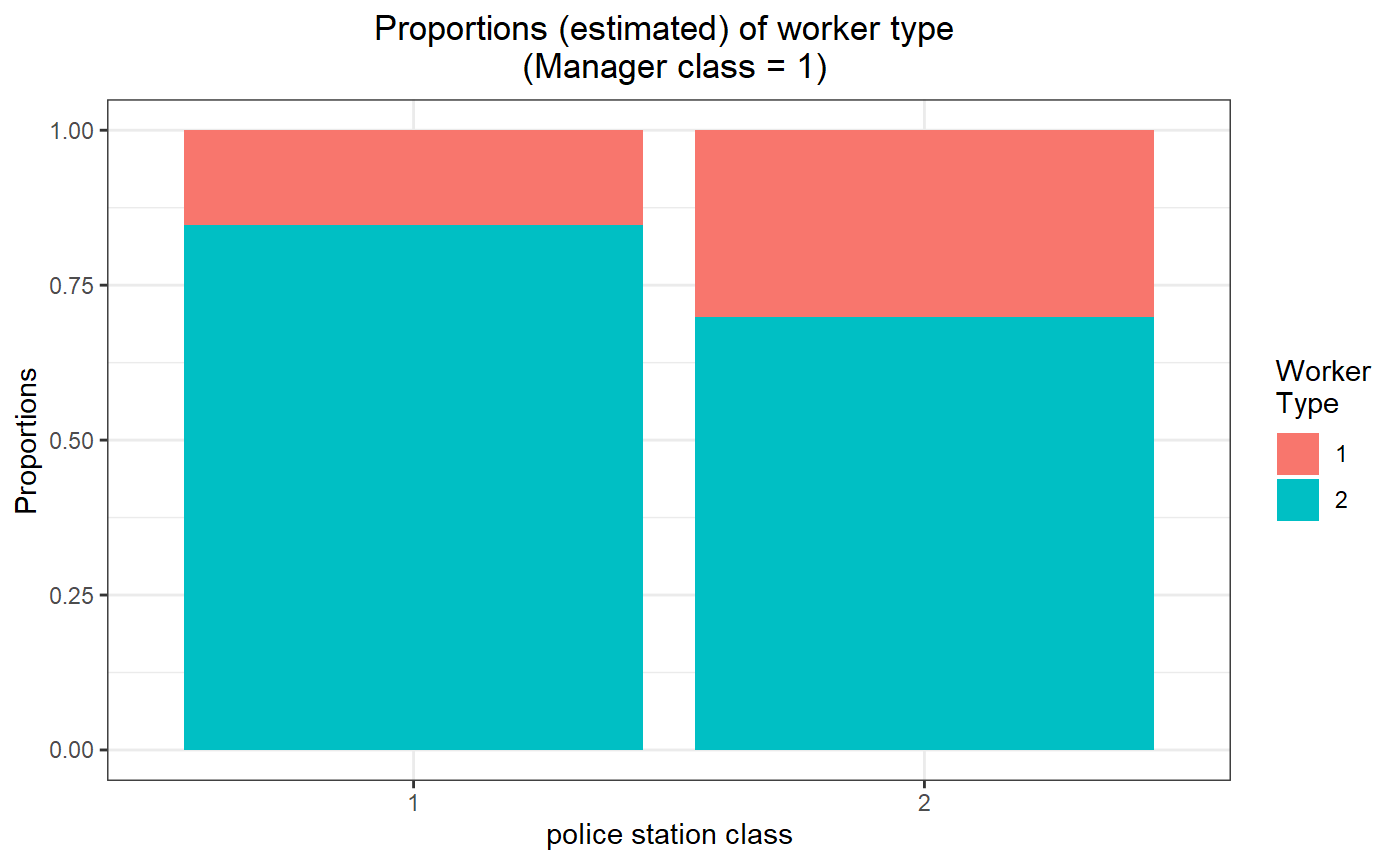
# for manager class 2
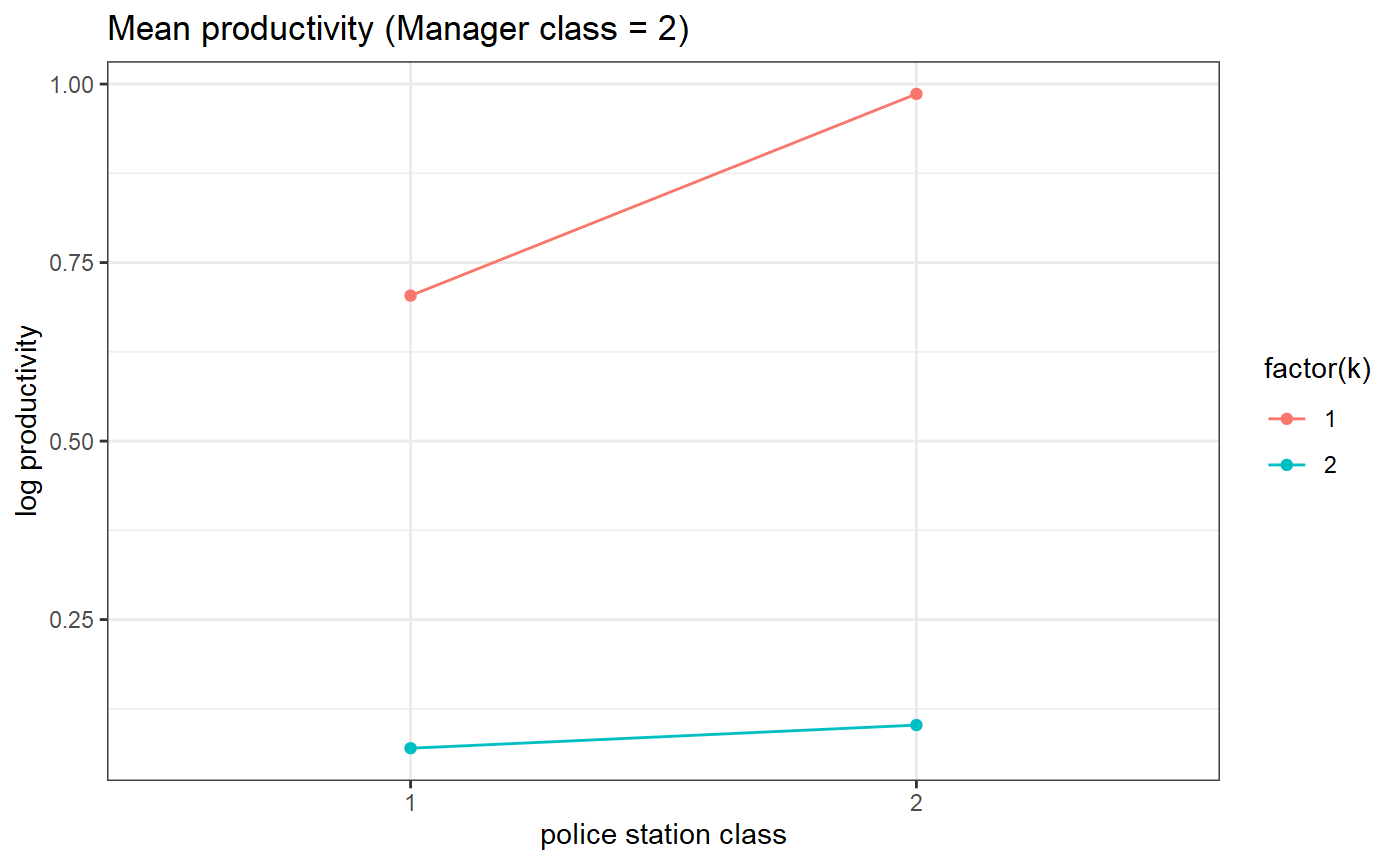
threeSided.proportion.plot(my_model_test_cluster, m=2)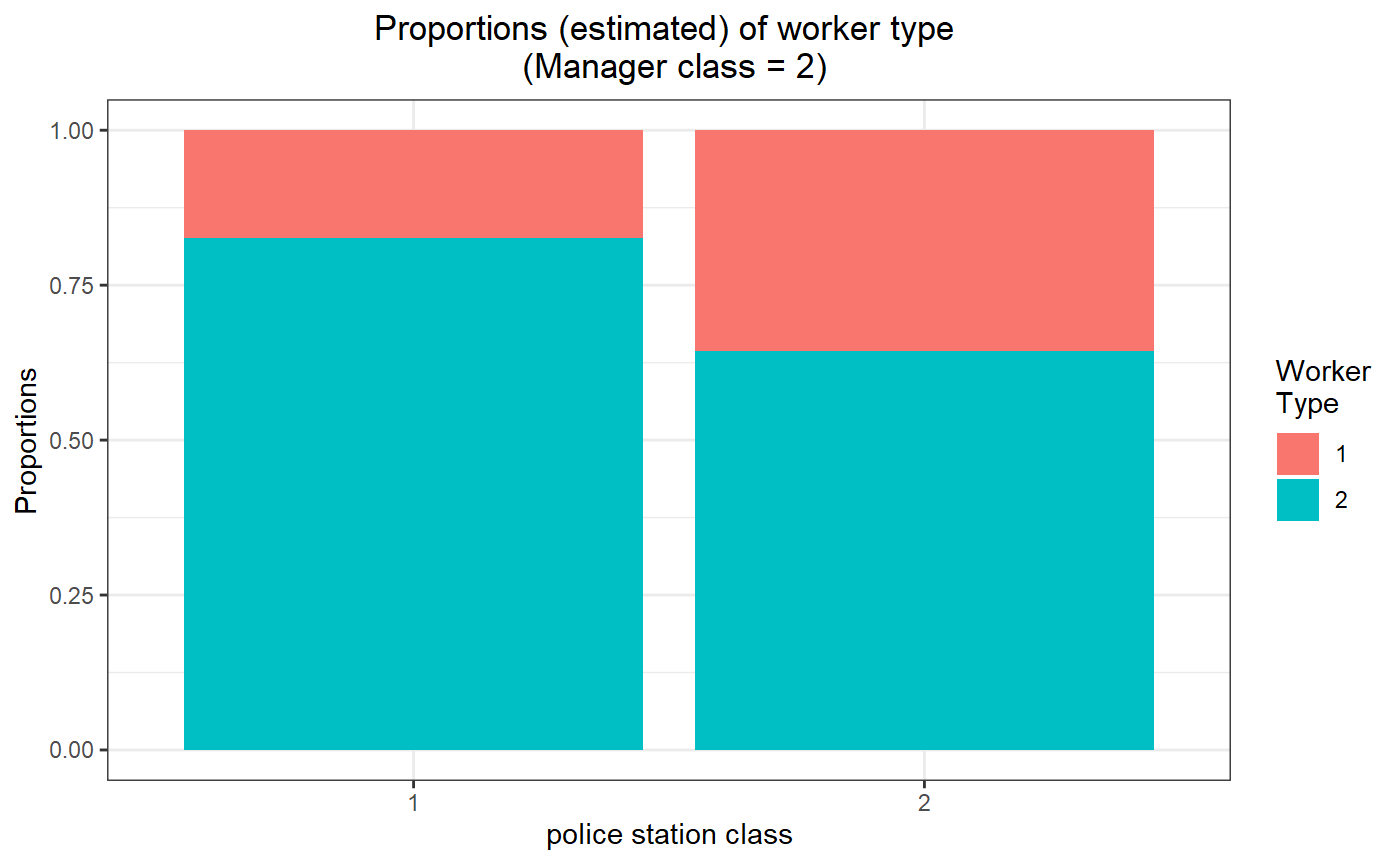 # Estimated parameters (means)
# Estimated parameters (means)
# for manager class 1
threeSided.means.plot(my_model_test_cluster, m=1)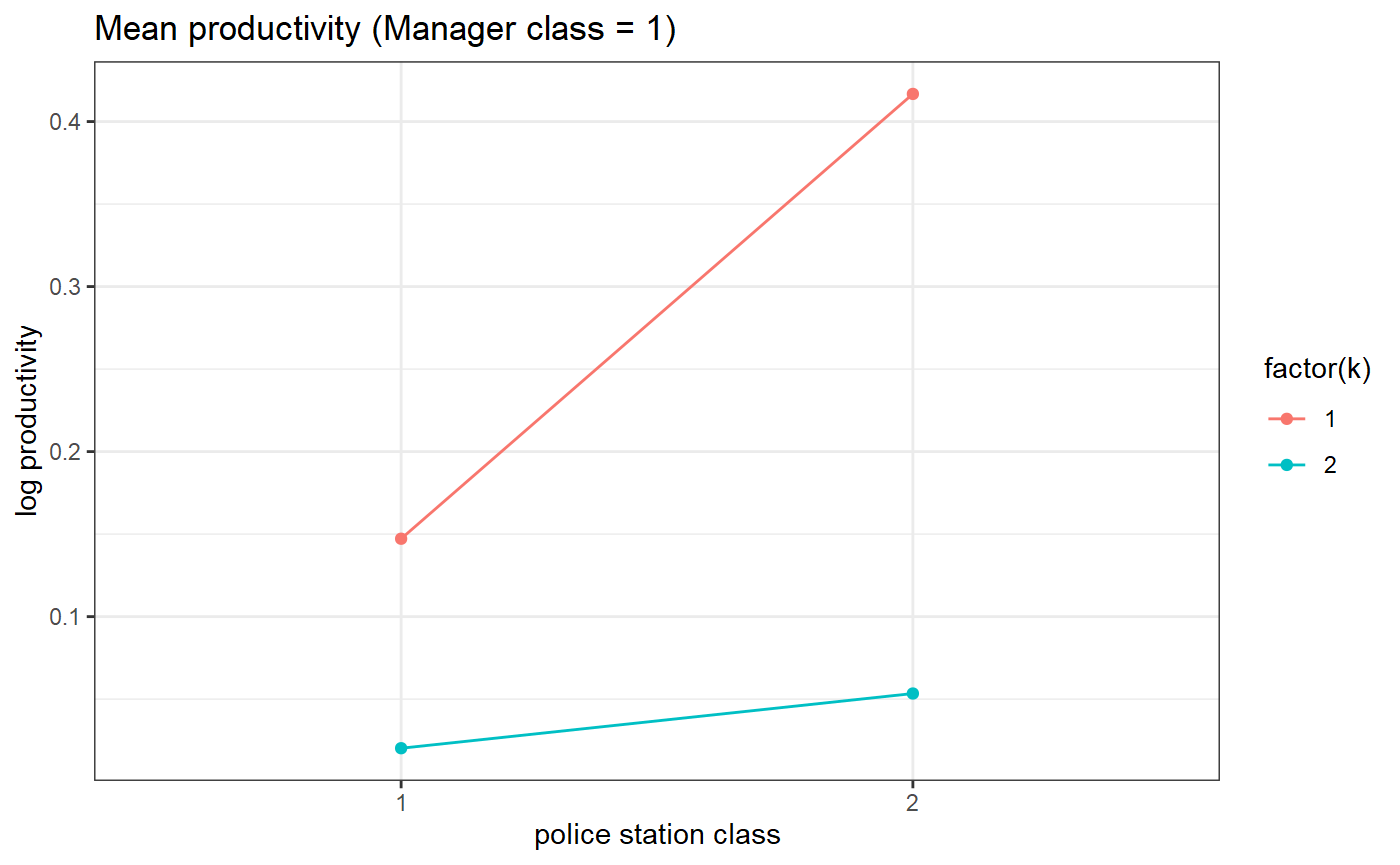
# for manager class 2
threeSided.means.plot(my_model_test_cluster, m=2)